import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm, LinearSegmentedColormap, Normalize, SymLogNorm
from matplotlib import cm as colormap
from matplotlib import animation, collections
import time
from string import ascii_lowercase
# SimPEG, discretize
import discretize
from discretize import utils
from simpeg.electromagnetics import time_domain as tdem
from simpeg.electromagnetics import resistivity as dc
from simpeg.utils import mkvc, plot_1d_layer_model
from simpeg import (
maps,
data,
data_misfit,
inverse_problem,
regularization,
optimization,
directives,
inversion,
utils,
Report
)from simpeg import electromagneticsSolver = utils.solver_utils.get_default_solver()csx = 100
csz = 50
scale = 1400
ncx = int(np.ceil(scale/csx))
ncz = int(np.ceil(scale/csx))
nca = 1
npadx = 12
npadz = 14
pf = 1.45
mesh = discretize.TensorMesh(
[
[(csx, npadx, -pf), (csx, ncx), (csx, npadx, pf)],
[(csx, npadx, -pf), (csx, ncx), (csx, npadx, pf)],
[(csz, npadz, -pf), (csz, ncz+nca), (csz, npadz, pf)]
],
origin = "CC0"
)
mesh.origin = mesh.origin + np.r_[0, 0, -mesh.h[2][:npadz+ncz].sum()]
mesh.plot_grid()<Axes3D: xlabel='x1', ylabel='x2', zlabel='x3'>
mesh
TensorMesh: 62,092 cells
MESH EXTENT CELL WIDTH FACTOR
dir nC min max min max max
--- --- --------------------------- ------------------ ------
x 38 -28,211.51 28,211.51 100.00 8,638.06 1.45
y 38 -28,211.51 28,211.51 100.00 8,638.06 1.45
z 43 -29,799.10 29,149.10 50.00 9,080.76 1.45
rho_back = 1000
rho_layer = 20
rho_air = 1e8
slope_layer = -0.2
layer_b = -350
t_layer = 200
def z_layer_top(y):
return np.min([np.zeros_like(y), slope_layer * y + layer_b], axis=0)
def z_layer_bottom(y):
return np.min([np.zeros_like(y), z_layer_top(y) - t_layer], axis=0)
wire_length = 400
def diffusion_distance(sigma, t):
return 1260 * np.sqrt(t/sigma)rx_times = np.logspace(np.log10(1e-6), np.log10(8e-3), 20)
diffusion_distance(1./rho_back, 1e-3)np.float64(1260.0)resistivity_model = rho_air * np.ones(mesh.n_cells)
resistivity_model[mesh.cell_centers[:, 2] < 0] = rho_back
halfspace = resistivity_model.copy()
layer_x = np.r_[-600, 600]
inds_z_layer = (
(mesh.cell_centers[:, 0] >= layer_x.min()) &
(mesh.cell_centers[:, 0] <= layer_x.max()) &
(mesh.cell_centers[:, 2] >= z_layer_bottom(mesh.cell_centers[:, 0])) &
(mesh.cell_centers[:, 2] <= z_layer_top(mesh.cell_centers[:, 0]))
)
resistivity_model[inds_z_layer] = rho_layer
models = {
"target": resistivity_model,
"halfspace": halfspace
}np.unique(resistivity_model)array([2.e+01, 1.e+03, 1.e+08])fig, ax = plt.subplots(1, 1, figsize=(8, 4))
xlim = 800*np.r_[-1, 1]
zlim = np.r_[-1200, 200]
key = "target"
mesh.plot_slice(
1./models[key], ax=ax,
pcolor_opts={"norm":LogNorm(vmin=1./rho_air*1000, vmax=1./rho_layer)},
normal="Y",
grid=True,
grid_opts={"color":"k", "lw": 0.5}
)
ax.set_xlim(xlim)
ax.set_ylim(zlim)
ax.set_aspect(1)
plt.tight_layout()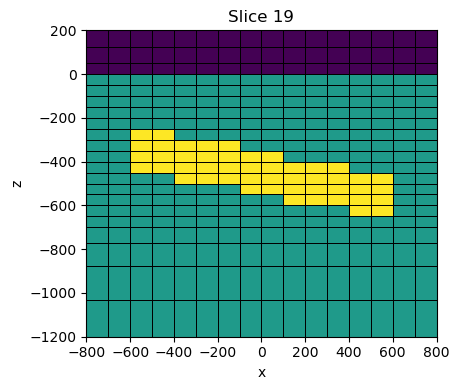
simulation¶
time_steps = [
# (1e-4, np.floor(waveform.peak_time/1e-4)),
# (1e-5, np.floor((waveform.off_time-waveform.peak_time)/1e-5)),
(1e-6, 40), (3e-6, 40), (1e-5, 40), (3e-5, 50), #(1e-4, 5), #(3e-4, 10)
]
rx_times = np.cumsum(np.hstack([np.r_[0], discretize.utils.unpack_widths(time_steps)]))
rx_times.max()*1e3np.float64(2.060000000000003)fig, ax = plt.subplots(1, 1, figsize=(4, 1.5), dpi=150)
x = np.linspace(-rx_times.max()/10, rx_times.max()*1.05, 1000)
waveform = np.zeros_like(x)
waveform[x < 0] = 1
ax.plot(x*1e3, waveform, color="k")
ax.grid()
rx_times_plot = 1
ax.plot(rx_times[::rx_times_plot]*1e3, np.zeros_like(rx_times[::rx_times_plot]), "|", markeredgewidth=0.75, ms=10, color="C3", label="simulation times")
ax.set_ylim(np.r_[-0.25, 1.25])
ax.legend()
ax.set_ylabel("TX current")
ax.set_xlabel("time (ms)")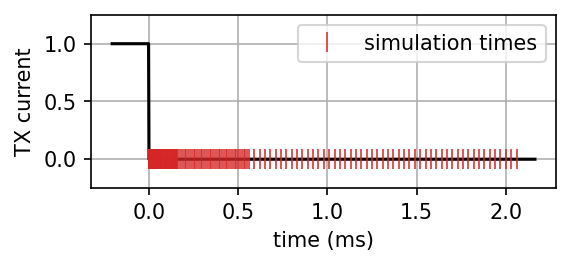
waveform=tdem.sources.StepOffWaveform()
rx_y = 500
rx_height = 0
rx_z = tdem.receivers.PointMagneticFluxTimeDerivative(
np.r_[0, rx_y, rx_height], times=rx_times, orientation="z"
)
loop_src = tdem.sources.CircularLoop(
radius = wire_length/2,
receiver_list=[rx_z], waveform=waveform, moment=-1
# srcType="inductive"
)
# loop_src = tdem.sources.LineCurrent(
# location = np.array([
# [-wire_length/2, -wire_length/2, 0],
# [-wire_length/2, wire_length/2, 0],
# [wire_length/2, wire_length/2, 0],
# [wire_length/2, -wire_length/2, 0],
# [-wire_length/2, -wire_length/2, 0],
# ]),
# receiver_list=[rx_z], waveform=waveform,
# srcType="inductive"
# )
wire_src = tdem.sources.LineCurrent(
location=np.array([
[0, -wire_length, 0],
[0, wire_length, 0],
]),
receiver_list=[rx_z], waveform=waveform,
srcType="galvanic"
)
survey = tdem.Survey([loop_src]) #, wire_src])sim = tdem.simulation.Simulation3DElectricField(
mesh=mesh, survey=survey, time_steps=time_steps, solver=Solver,
rhoMap=maps.IdentityMap(mesh)
)fields = {}
dpred = {}
for key, val in models.items():
t = time.time()
fields[key] = sim.fields(val)
print(f"done fields {key}... {time.time() - t: 1.2e}s")
dpred[key] = sim.dpred(val, f=fields[key])
print(f"done dpred {key}... {time.time() - t: 1.2e}s")
/Users/lindseyjh/miniforge3/envs/py311/lib/python3.11/site-packages/pymatsolver/solvers.py:415: FutureWarning: In Future pymatsolver v0.4.0, passing a vector of shape (n, 1) to the solve method will return an array with shape (n, 1), instead of always returning a flattened array. This is to be consistent with numpy.linalg.solve broadcasting.
return self.solve(val)
done fields target... 5.62e+01s
done dpred target... 5.66e+01s
done fields halfspace... 5.68e+01s
done dpred halfspace... 5.72e+01s
def plot_data(
ti=None, key="target",src="loop", ax=None, xlim=np.r_[1e-2, 2], ylim=np.r_[1e-10, 1e-4]
):
if ax is None:
fig, ax = plt.subplots(1, 1, figsize=(4.5, 3.5))
def plot_loglog(vals, color, label):
neg_inds = vals < 1e-9
neg_inds[0] = False
ax.loglog(rx_times[neg_inds]*1e3, -vals[neg_inds], color, label=label)
ax.loglog(rx_times[~neg_inds]*1e3, vals[~neg_inds], "--" + color)
for i, key in enumerate(["halfspace", "target"]):
if src == "loop":
dpred_vals = dpred[key][:len(rx_times)]
elif src == "wire":
dpred_vals = dpred[key][len(rx_times):]
plot_loglog(dpred_vals, f"C{i}", key)
if ti is not None:
ax.plot(sim.times[ti]*1e3*np.r_[1, 1], ylim, color="k", lw=1)
# ax.plot(sim.times[ti]*1e3, -dpred["target"][ti], ".k", ms=8)
time_label = (sim.times[ti]-waveform.off_time)*1e3
if time_label > 0.99:
ax.text(0.9, ylim.min()*1.2, f"t={time_label:1.1f} ms", fontsize=14, ha="right")
else:
ax.text(0.9, ylim.min()*1.2, f"t={time_label:1.2f} ms", fontsize=14, ha="right")
ax.set_xlim(xlim)
ax.set_ylim(ylim)
ax.legend()
ax.set_xlabel("time (ms)")
ax.set_ylabel("db/dt")
ax.grid(alpha=0.7)
return axax = plot_data(1, src="loop")
# ax.set_ylim(1e-15, 1)
# ax.set_xlim(1e-3, 1e1)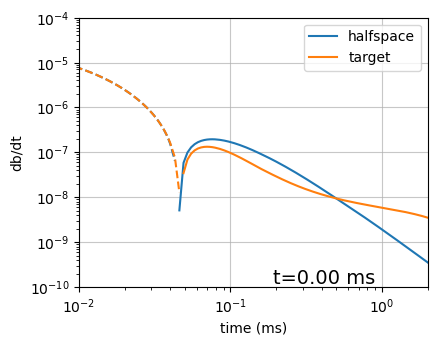
def plot_model(
ti, key = "target", ax=None, xlim=1200*np.r_[-1, 1], zlim=np.r_[-1200, 200],
colorbar=False, ylabels=True, vmax=None, cax=None, src="loop"
):
if ax is None:
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
cb = plt.colorbar(mesh.plot_slice(
1./models[key], ax=ax,
pcolor_opts={"norm":LogNorm(vmin=1./rho_air*1000, vmax=1./rho_layer)},
normal="Y",
grid=True,
grid_opts={"color":[0.5, 0.5, 0.5], "lw": 0.5}
)[0], ax=ax, cax=cax)
ax.set_xlim(xlim)
ax.set_ylim(zlim)
ax.set_aspect(1)
plt.tight_layout()
cb.set_label("conductivity (S/m)")
if src == "loop":
ax.plot(-wire_length/2, 0, "wo", ms=6)
ax.plot(wire_length/2, 0, "wo", ms=6)
elif src == "wire":
ax.plot(0, 0, "wo", ms=6)
ax.plot(rx_y, rx_height, "ws", ms=6)
# ax.plot(tx_radius*np.r_[-1, 1], tx_height*np.r_[1, 1], color="C1", lw=2)
ax.text(-wire_length/2, 0+40, f"TX", fontsize=14, ha="center", color="w")
ax.text(rx_y, rx_height+40, f"RX", fontsize=14, ha="center", color="w")
ax.set_title("")
ax.text(xlim.min() + 20, zlim.min() + 100, f"{rho_back} $\Omega$m", fontsize=14)
if key =="target":
ax.text(layer_x.min() + 20, -400, f"{rho_layer} $\Omega$m", fontsize=14)
ax.text(xlim.min() + 20, 2, f"{rho_air:1.0e} $\Omega$m", color="w", fontsize=14)
# ax.text(0, target_z, f"{1/sigma_target:1.0f} $\Omega$m", fontsize=14, ha="center", va="center")
ax.set_xlim(xlim)
ax.set_ylim(zlim)
ax.set_aspect(1)
plot_model(0)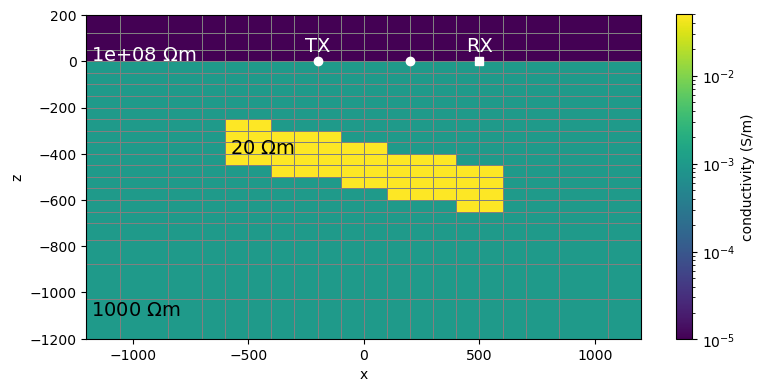
fig, ax = plt.subplots(2, 1, figsize = (7, 7.5), dpi=150)
plot_data(ax=ax[0])
plot_model(ax=ax[1], ti=0)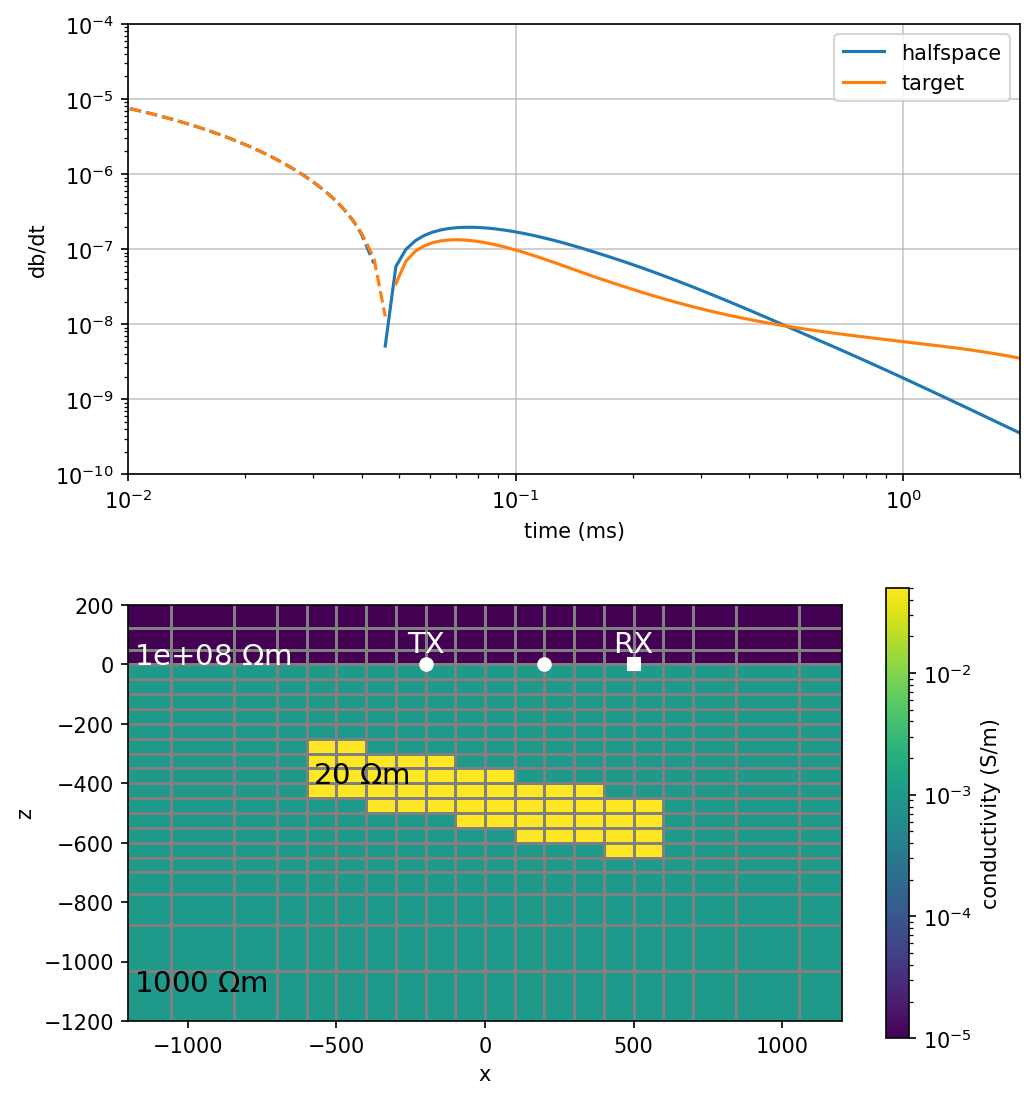
def plot_current_density(
ti, key = "target", src="loop", ax=None, xlim=1200*np.r_[-1, 1], zlim=np.r_[-1200, 200],
colorbar=False, ylabels=True, vmax=None, cax=None
):
if ax is None:
fig, ax = plt.subplots(1, 1, figsize=(4, 4))
if src == "loop":
jplt = (mesh.average_edge_y_to_cell * np.squeeze(fields[key][loop_src, "j", ti])[mesh.n_edges_x : np.sum(mesh.n_edges_per_direction[:2])])
# if vmax is not None:
vmax = np.max(np.abs(jplt))
pcolor_opts={"norm":Normalize(vmin=-vmax, vmax=vmax), "cmap":"coolwarm"}
# else:
# pcolor_opts = {"cmap":"coolwarm"}
out = mesh.plot_slice(
jplt, ax=ax,
pcolor_opts=pcolor_opts,
normal="y"
)
# ax.plot(-wire_length/2, 0, "ko", ms=2)
# ax.plot(wire_length/2, 0, "ko", ms=2)
# ax.plot(0, rx_height, "ks", ms=2)
# ax.plot(0, 0, "ko", ms=2)
elif src == "wire":
src_ind = wire_src
ax.set_xlim(xlim)
ax.set_ylim(zlim)
ax.set_aspect(1)
if colorbar is True:
cb = plt.colorbar(out[0], ax=ax, cax=cax)
ticks = cb.get_ticks()
cb.formatter.set_scientific(True)
cb.formatter.set_powerlimits((0,0))
cb.set_ticks([ticks[1], 0, ticks[-2]])
cb.update_ticks()
cb.set_label("current density")
if ylabels is False:
ax.set_ylabel("")
ax.set_yticklabels("")
ax.set_title("")
return cbdef plot_dbdt(
ti, key = "target", ax=None, xlim=1200*np.r_[-1, 1], zlim=np.r_[-1200, 200],
colorbar=False, ylabels=True, vmax=1e-2, vmin=3e-10, cax=None
):
if ax is None:
fig, ax = plt.subplots(1, 1)
dbdtplt = -1 * mesh.average_face_to_cell_vector * fields[key][:, "dbdt", ti]
vmin = np.max([vmin, 1e-4*np.max(np.abs(dbdtplt))])
out = mesh.plot_slice(
dbdtplt, "CCv", view="vec", ax=ax,
pcolor_opts={"norm":LogNorm(vmin=vmin, vmax=vmax)},
range_x=xlim,
range_y=zlim,
stream_threshold=vmin,
normal="y"
)
# ax.plot(np.r_[0], tx_height, "ko", ms=2)
# ax.plot(-wire_length/2, 0, "ko", ms=2)
# ax.plot(wire_length/2, 0, "ko", ms=2)
ax.plot(rx_y, rx_height, "ws", ms=6)
ax.set_title("")
ax.set_xlim(xlim)
ax.set_ylim(zlim)
ax.set_aspect(1)
if ylabels is False:
ax.set_ylabel("")
ax.set_yticklabels("")
if colorbar is True:
cb = plt.colorbar(out[0], ax=ax, cax=cax)
cb.set_label("db/dt")plot_dbdt(120)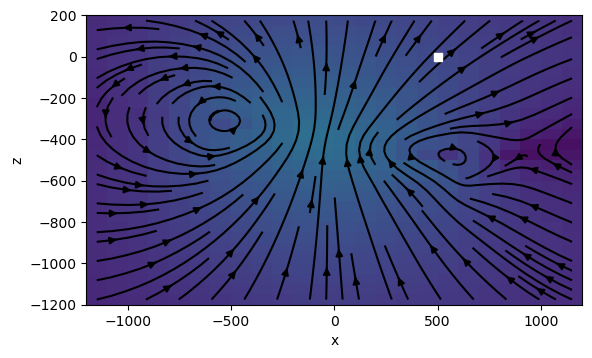
fig, ax = plt.subplots(2, 2, figsize=(14, 6.5), dpi=150)
ti = 55
plot_model(ti, ax=ax[0, 0])
plot_data(ti, ax=ax[0, 1])
plot_current_density(ti, ax=ax[1, 0], colorbar=True)
plot_dbdt(ti, ax=ax[1, 1], colorbar=True)
ax[0, 1].set_aspect(0.2)
# plt.tight_layout()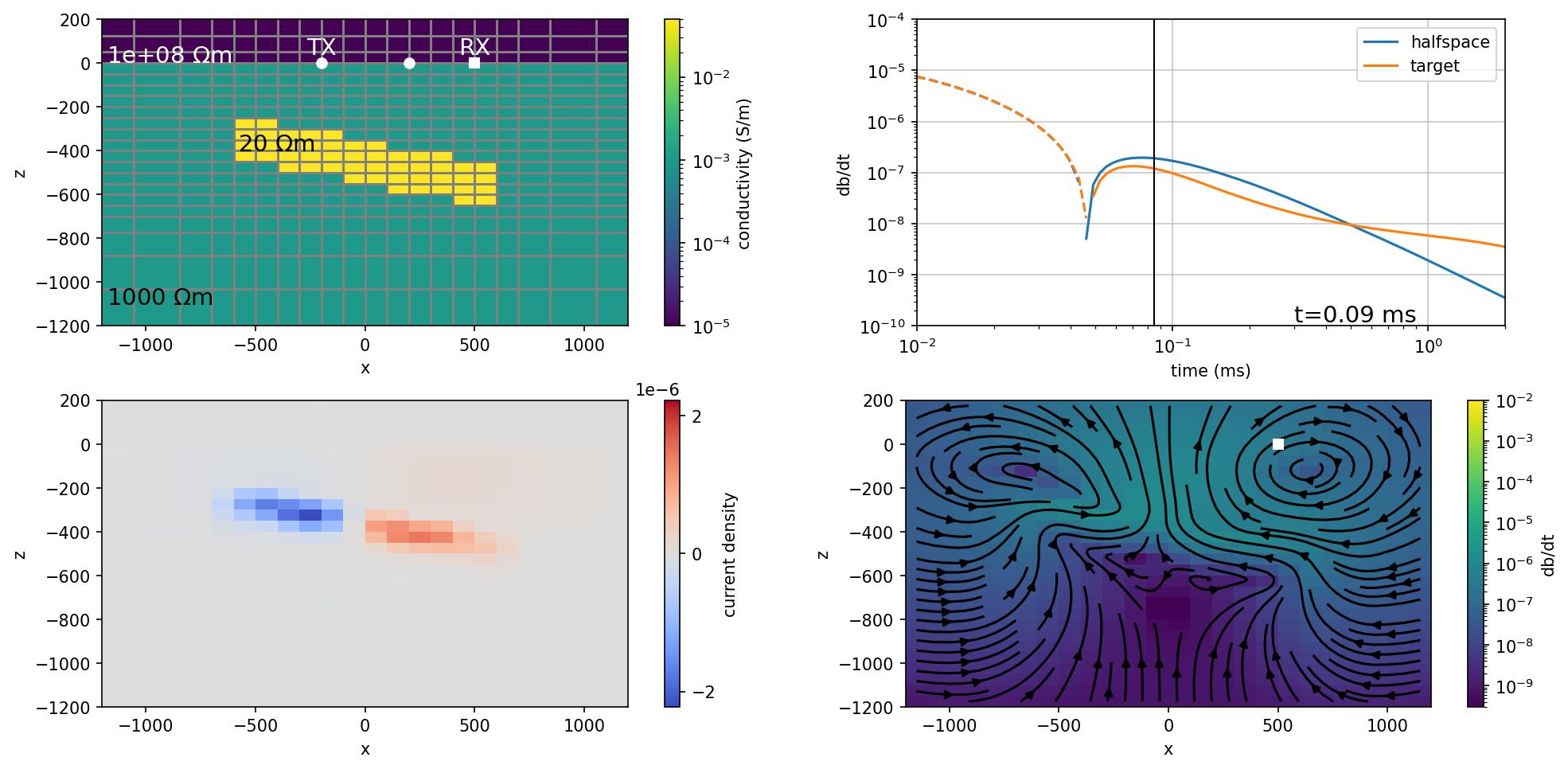
times = np.where(sim.times*1e3 < 1)[0]
ims = []
fig, ax = plt.subplots(2, 2, figsize=(14, 6.5), dpi=250)
def plotme(ti):
for a in ax.flatten():
a.clear()
if len(fig.get_axes()) > 4:
axes = fig.get_axes()
cax = axes[-3:]
else:
axes = ax.flatten()
cax = None
plot_model(ti, ax=axes[0], colorbar=True, cax=cax[-3] if cax is not None else None)
plot_data(ti, ax=axes[1])
plot_current_density(ti, ax=axes[2], colorbar=True, cax=cax[-2] if cax is not None else None)
plot_dbdt(ti, ax=axes[3], colorbar=True, cax=cax[-1] if cax is not None else None)
axes[1].set_aspect(0.2)
# plt.tight_layout()
return ax
out = plotme(0)
def init():
[o.set_array(None) for o in out if isinstance(o, collections.QuadMesh)]
return
def update(t):
for a in ax.flatten():
a.clear()
return plotme(t)
ani = animation.FuncAnimation(fig, update, times, init_func=init, blit=False)
ani.save(
f"./dipping_inductive_video2{key}.mp4", writer="ffmpeg", fps=5, bitrate=0,
metadata={"title":f"TDEM {key} currents", "artist":"Lindsey Heagy"}
)
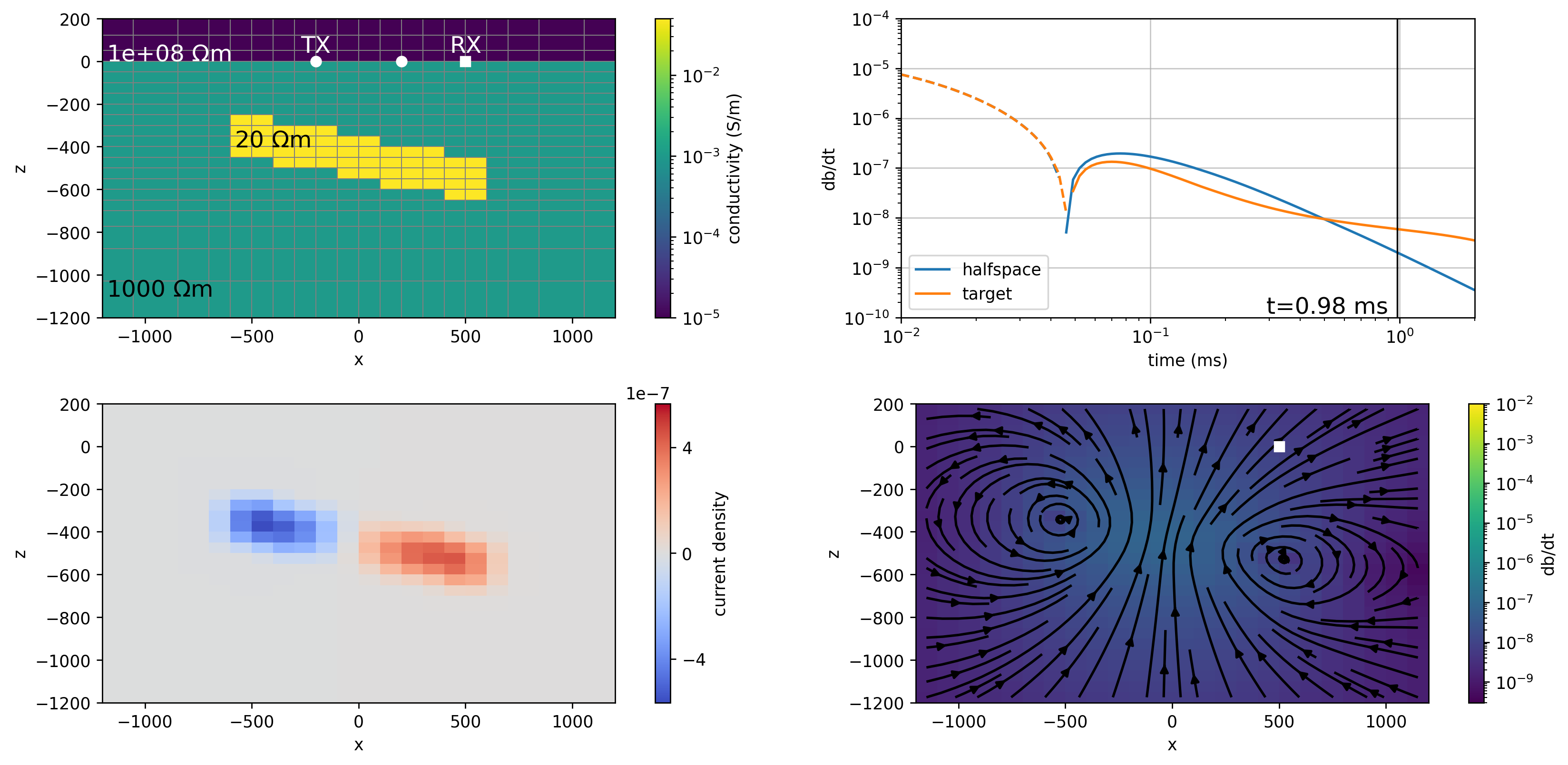
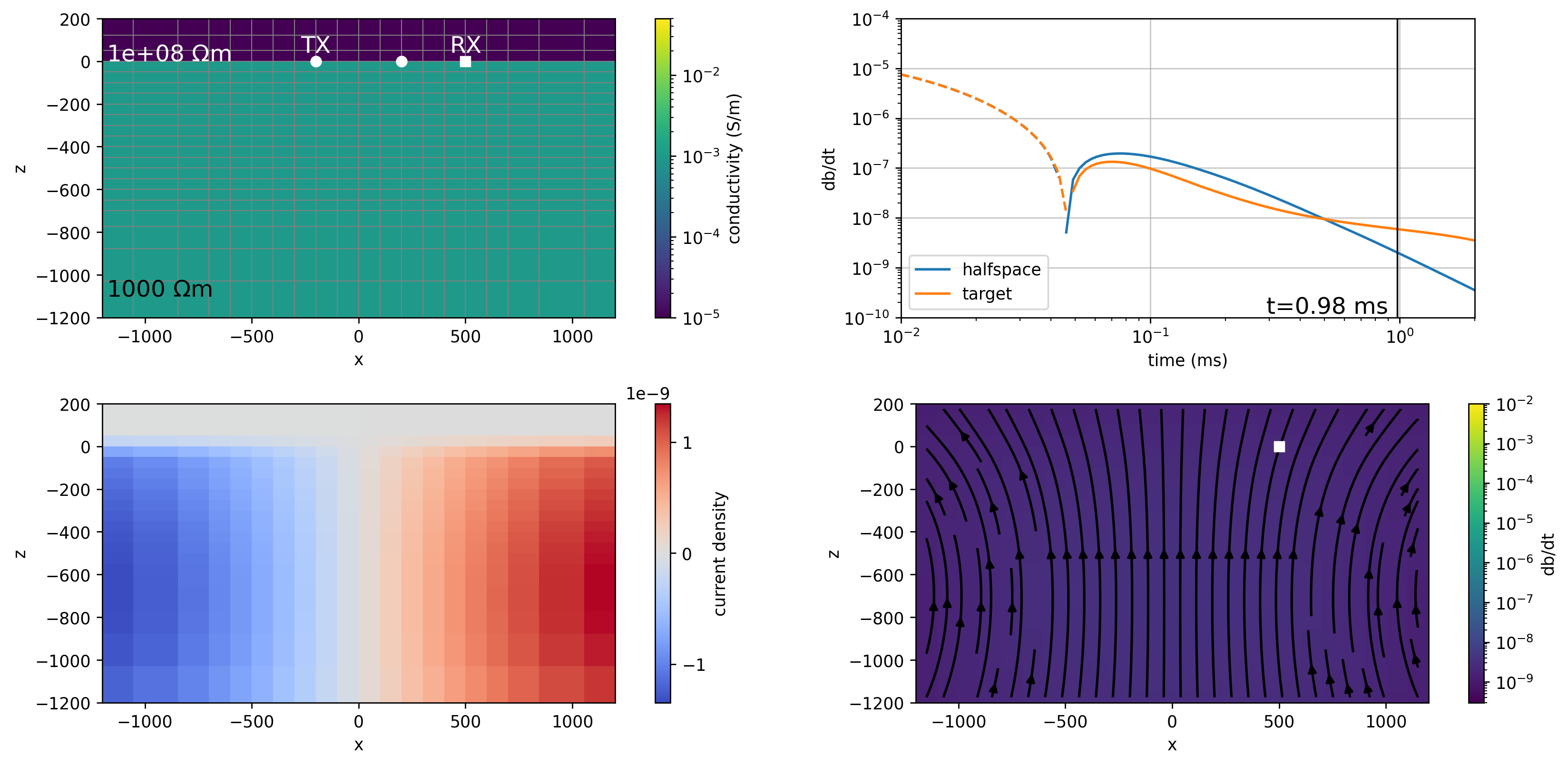
times = np.where(sim.times*1e3 < 1)[0]
ims = []
fig, ax = plt.subplots(2, 2, figsize=(14, 6.5), dpi=250)
def plotme(ti):
for a in ax.flatten():
a.clear()
if len(fig.get_axes()) > 4:
axes = fig.get_axes()
cax = axes[-3:]
else:
axes = ax.flatten()
cax = None
plot_model(ti, key="halfspace", ax=axes[0], colorbar=True, cax=cax[-3] if cax is not None else None)
plot_data(ti, ax=axes[1])
plot_current_density(ti, key="halfspace", ax=axes[2], colorbar=True, cax=cax[-2] if cax is not None else None)
plot_dbdt(ti, key="halfspace", ax=axes[3], colorbar=True, cax=cax[-1] if cax is not None else None)
axes[1].set_aspect(0.2)
# plt.tight_layout()
return ax
out = plotme(0)
def init():
[o.set_array(None) for o in out if isinstance(o, collections.QuadMesh)]
return
def update(t):
for a in ax.flatten():
a.clear()
return plotme(t)
ani = animation.FuncAnimation(fig, update, times, init_func=init, blit=False)
ani.save(
f"./halfspace_inductive_video2{key}.mp4", writer="ffmpeg", fps=3, bitrate=0,
metadata={"title":f"TDEM {key} currents", "artist":"Lindsey Heagy"}
)
Report()
--------------------------------------------------------------------------------
Date: Sat Jan 25 12:22:51 2025 PST
OS : Darwin (macOS 13.6.6)
CPU(s) : 12
Machine : arm64
Architecture : 64bit
RAM : 64.0 GiB
Environment : Jupyter
File system : apfs
Python 3.11.10 | packaged by conda-forge | (main, Oct 16 2024, 01:26:25)
[Clang 17.0.6 ]
simpeg : 0.22.2.dev41+ge21040475
discretize : 0.11.0
pymatsolver : 0.3.1
numpy : 2.0.0
scipy : 1.14.1
matplotlib : 3.9.2
geoana : 0.7.2
libdlf : 0.3.0
cython : 3.0.11
numba : 0.60.0
sklearn : 1.5.2
pandas : 2.2.3
sympy : 1.13.3
IPython : 8.29.0
ipywidgets : 8.1.5
plotly : 5.24.1
--------------------------------------------------------------------------------